Research
Topics
1. Development of methodology to efficiently synthesize biologically active compounds
2. Development of small molecules to analyze and control protein methylation
4. Alkyne-tag Raman Imaging (ATRI) ~Visualization of small molecules in live cells~
5. Methodologies for identification of binding proteins/sites of bioactive compounds.
1. Development of methodology to efficiently synthesize biologically active compounds
We are involved in the development of new reactions in order to efficiently synthesize biologically active molecules.
1) Providing access to a wide range of F-containing molecules
Fluorine-containing molecules have had a lot of interest as drugs and agrochemicals due to their extraordinary characteristics including improved pharmacokinetics and unique molecular interactions due to fluorine atom. In addition, they have attracted increasing attention as a probe molecule for MRI and PET imagings. Thus, a diverse array of fluorine-containing molecules and their efficient synthetic methods are on demand. In this context, our group has been working on development of new synthetic reactions for fluorine-containing molecules. In particular, recent our focus has been on development of electrophilic perfluoroalkylations in order to provide useful perfluoroalkyl group-containing building blocks. Reactivity control of perfluoroalkylating reagents by the aid of catalyst or substrate design realized efficient and practical perfluoroalkylation reactions, which greatly extended the variety of available perfluoroalkyalted compounds. Furthermore, complementary experimental and theoretical mechanistic studies gave important scientific insights for designing new reactions in future.
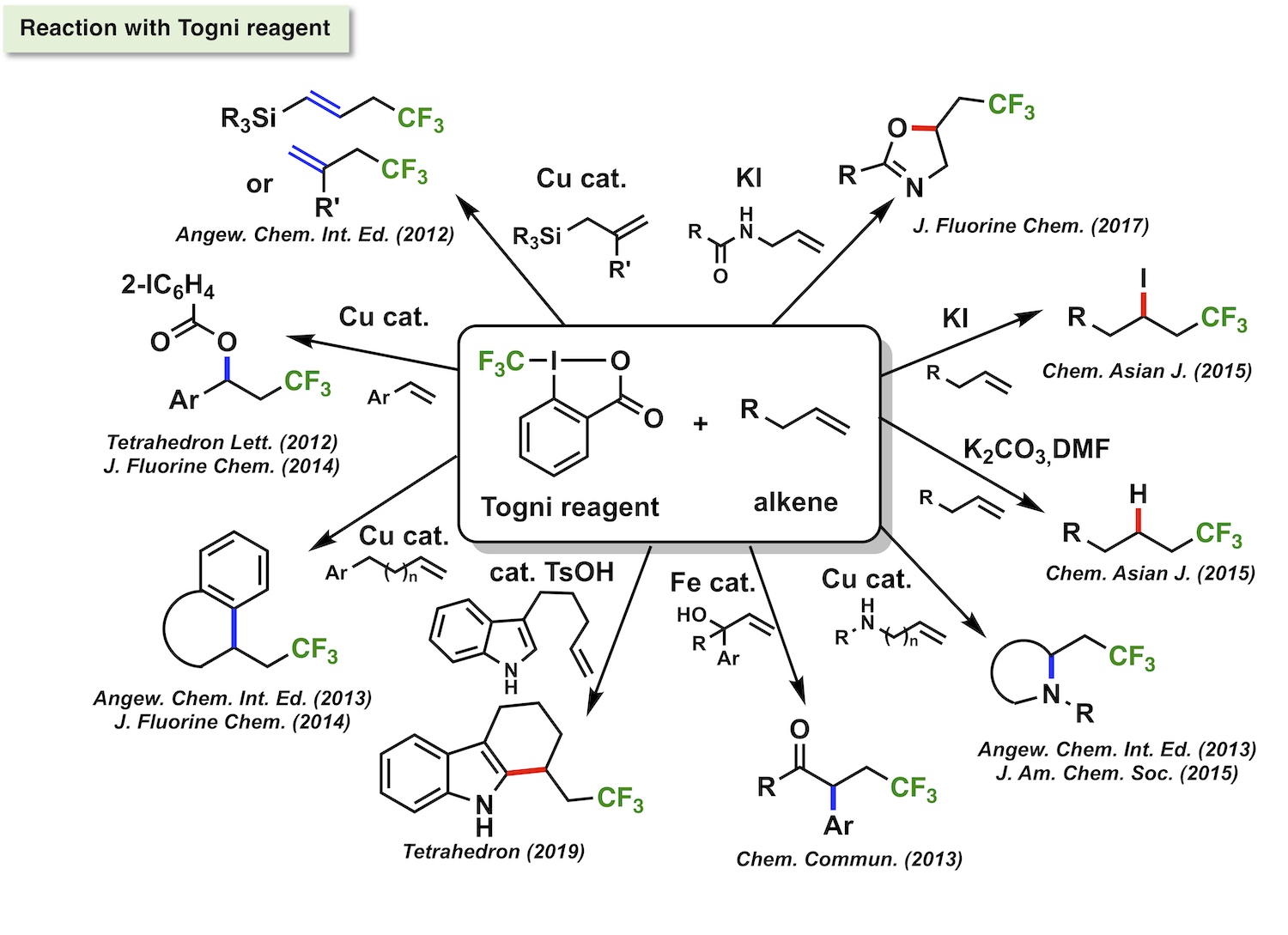
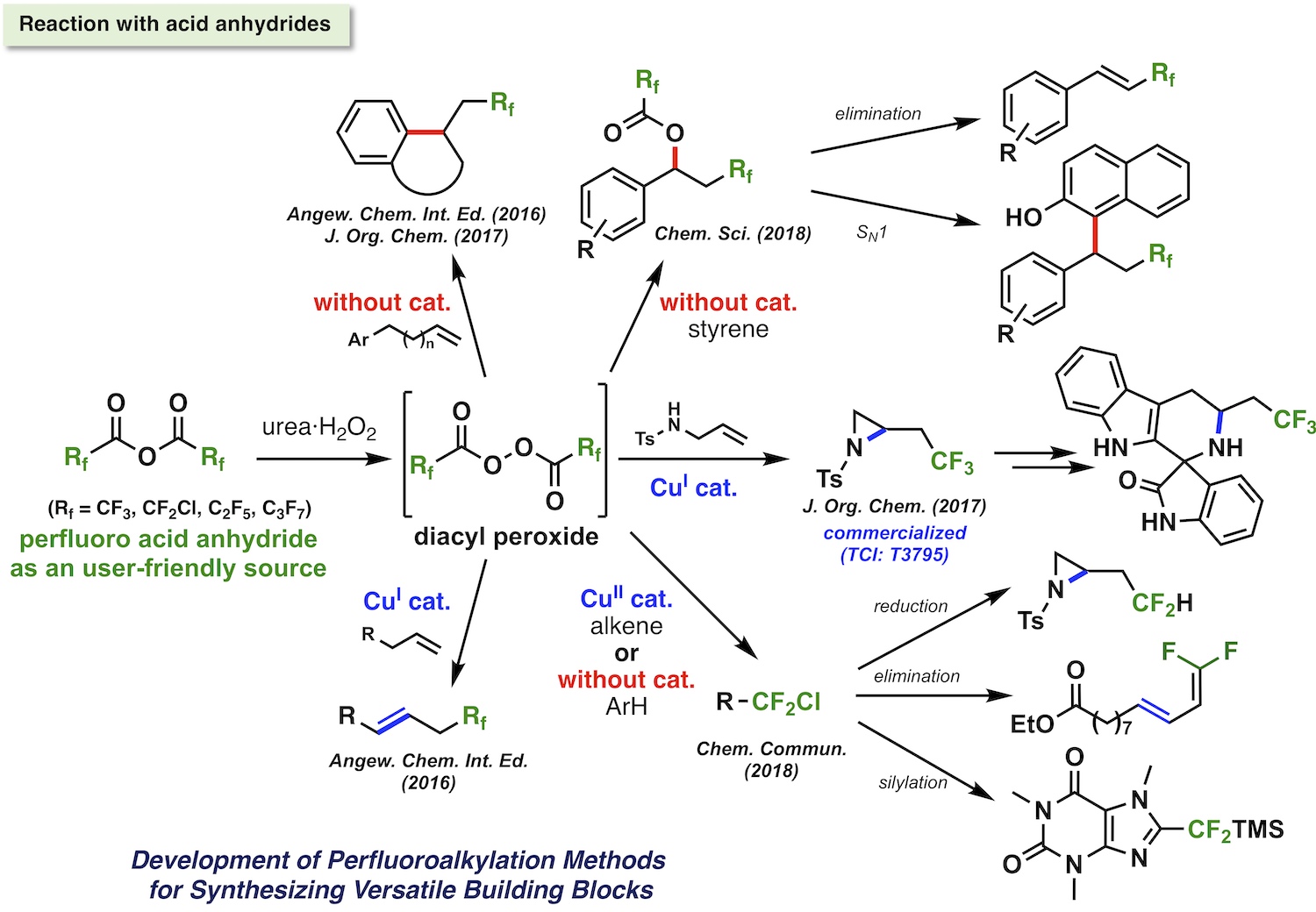
Press release
- RIKEN RESEARCH Research Highlights (August 19, 2016)
- Chem-Station (June 29, 2016)
- RIKEN RESEARCH Research Highlights (June 19, 2015)
- RIKEN RESEARCH Research Highlights (September 27, 2013)
- RIKEN RESEARCH Research Highlights (October 5, 2012)
2) Development of catalytic asymmetric reactions
The coming of the post-genome era has induced the need to develop biologically active compounds that can selectively control chiral target molecules such as proteins. In order to develop catalytic systems that can be applied to various asymmetric reactions, the development of catalytic reactions based on new concepts is significant. Our group has focused on the use of soft transition metals, such as palladium and nickel, towards the development of new catalytic complexes and reactions. These transition metal catalysts induce the formation of chiral enolates from carbonyl compounds in water or alcoholic solvents, enabling various asymmetric reactions under mild conditions. Based on this concept, we succeeded in the development of asymmetric carbon-carbon bond formation reactions, asymmetric fluorination reactions, cycloadditions and asymmetric cascade reactions, leading to the generation of various asymmetric molecules.
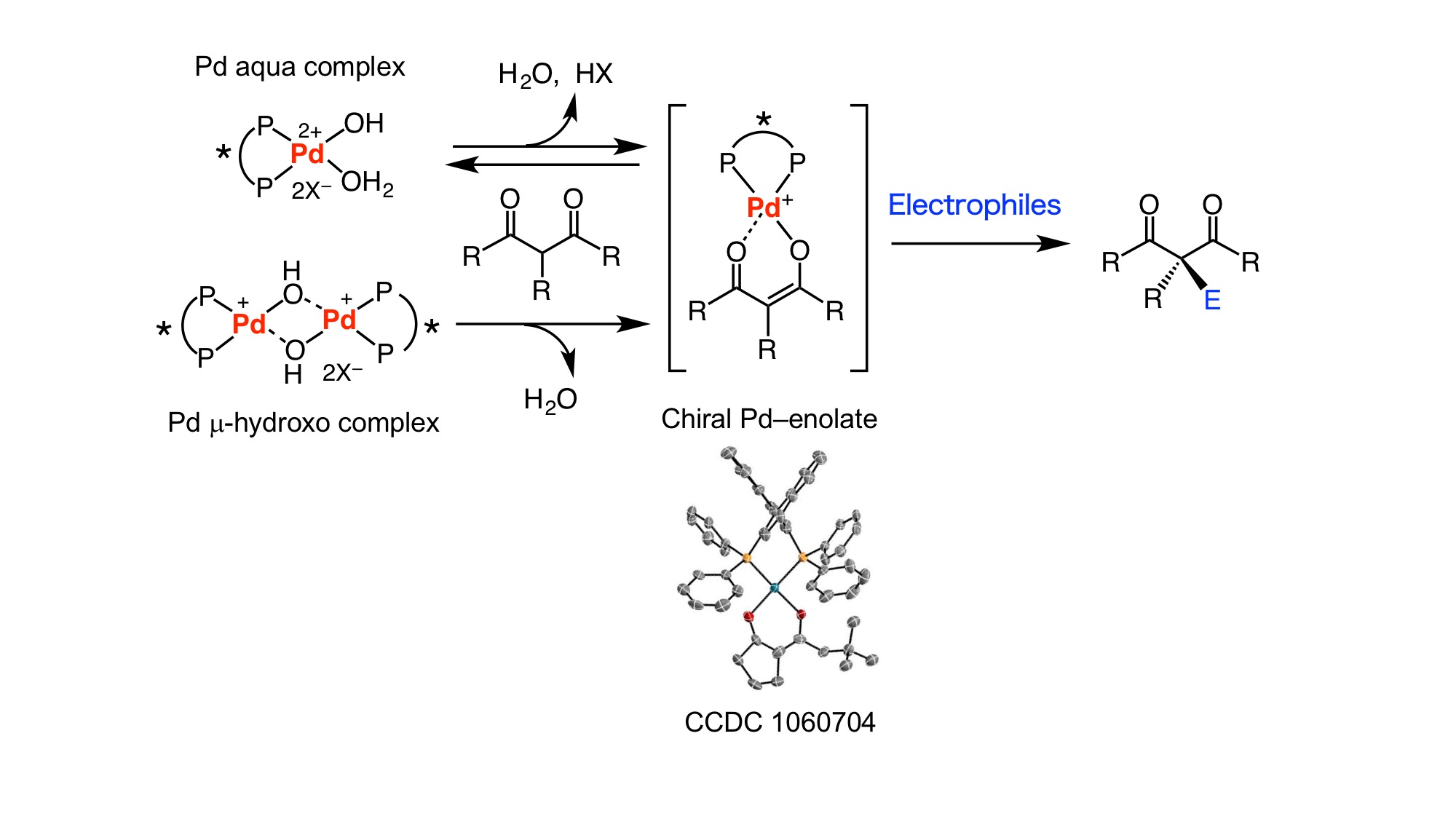
Press release
- RIKEN RESEARCH Research Highlights (September 28, 2012)
- RIKEN RESEARCH Research Highlights (June 4, 2010)
- RIKEN RESEARCH Research Highlights (September 19, 2008)
- RIKEN RESEARCH Research Highlights (September 7, 2007)
- RIKEN RESEARCH Research Highlights (November 24, 2006)
3) Development of metal complexes and their structural characterization
In general, optimization of the selectivity (enantio-, diastereo-, regio-, and chemoselectivity) of asymmetric catalysis relies on trial-and-error screening using the inherent stereochemistry embedded in “privileged” chiral templates. We aim to quantitatively characterize the bonding nature of the catalyst both in the solid- and solution states to explore the interplay of structure and selectivity in asymmetric acid-base catalysis. A successful example is shown below. Electron density distribution (EDD) analysis showed that the distortion around the Ni(II) centre makes the dz2 orbital partially ‘naked’, wherein the labile acetate ligand is coordinated with electrostatic interaction (in collaboration with Dr. Hashizume in RIKEN CEMS, Materials Characterization Support Team). The electron-deficient dz2 orbital and the acetate act together to deprotonate the α-ketoester, generating the (Λ)-Ni(II)–enolate. The solid and solution state analyses, together with theoretical calculations (in collaboration with Prof. Dr. Uchiyama and Dr. Muranaka in RIKEN CPR, Elements Chemistry Laboratory), strongly link the electronic structure of the centrochiral octahedral Ni(II) complex and its catalytic activity, depicting a cooperative mechanism of enolate binding and outer sphere hydrogen-bonding activation.
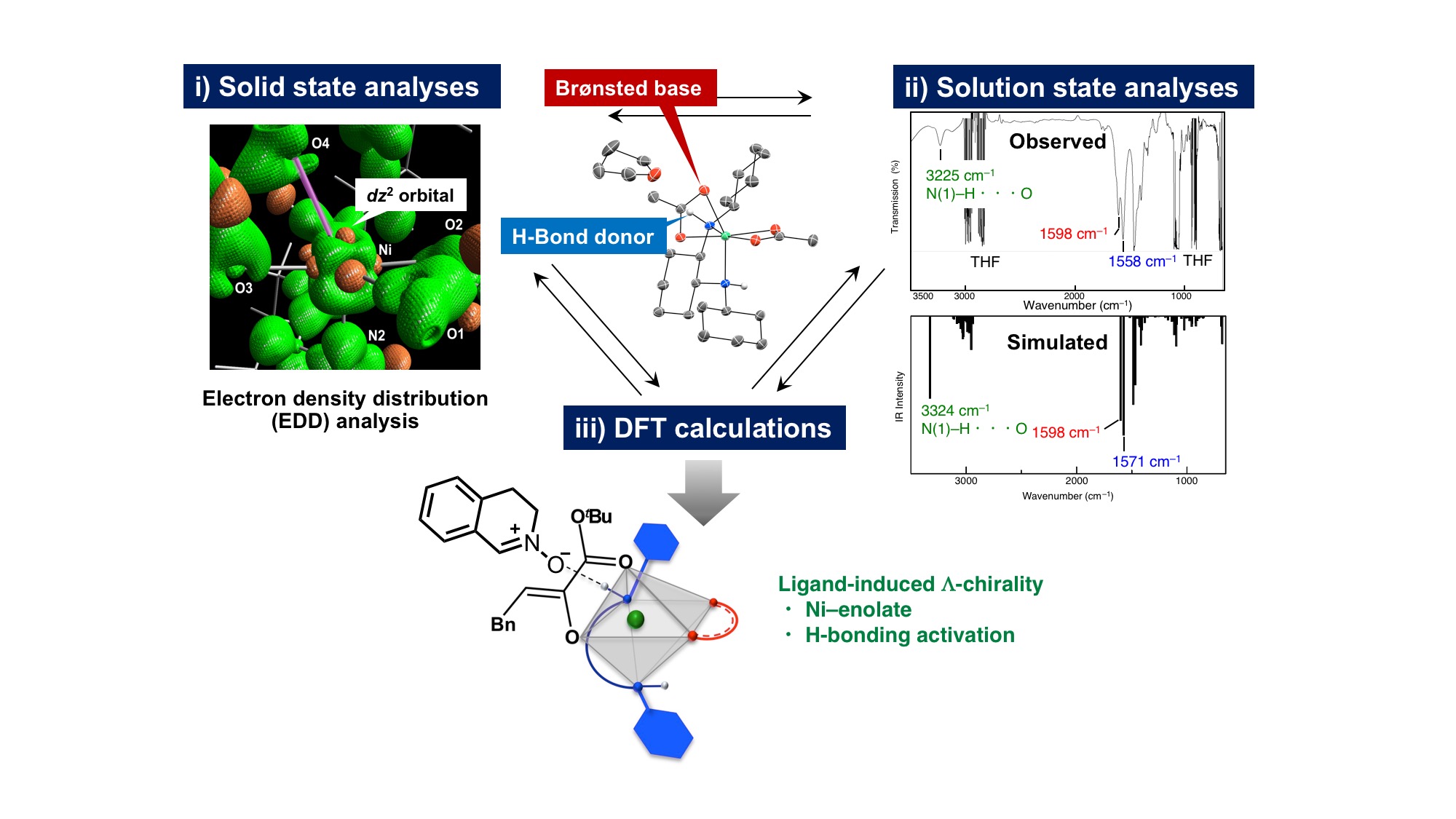
Press release
2. Development of small molecules to analyze and control protein methylation
Protein methyl transferases (PMTs) are one of the largest classes of epigenetic enzymes that participate in control of gene expression through methyl group transfer from S-adenosylmethionine (SAM, also called AdoMet) to their protein substrates. However, we lack a comprehensive methylome map of PMTs, their substrates, and their methylation sites. Considering that SAM is a common methyl donor in protein methylation, we have focused on SAM analogs for substrate-labeling. We identified that propargylic Se-adenosyl-L-selenomethionine (ProSeAM) is a useful tool for enriching the native methylome, which includes the following steps; (i) ProSeAM-mediated propargylation, (ii) Cu(I)-catalyzed alkyne-azide cycloaddition (CuAAC) to incorporate biotin and (iii) enrichment of the biotinylated substrates using streptavidin-immobilized beads. Specifically, we uncovered that the addition of a catalytic amount of exogenous recombinant PMT can enhance chemical modification with the ProSeAM. By comparing the enrichment values in the absence and presence of the exogenous PMT of interest, the numbers of substrate candidates picked up with the ProSeAM could be efficiently reduced. We also applied the developed chemical methylome analysis platform to evaluate the inhibitory activity of small molecules towards poorly characterized PMTs, facilitating to identify a new class of a non-histone arginine methyltransferase inhibitor.
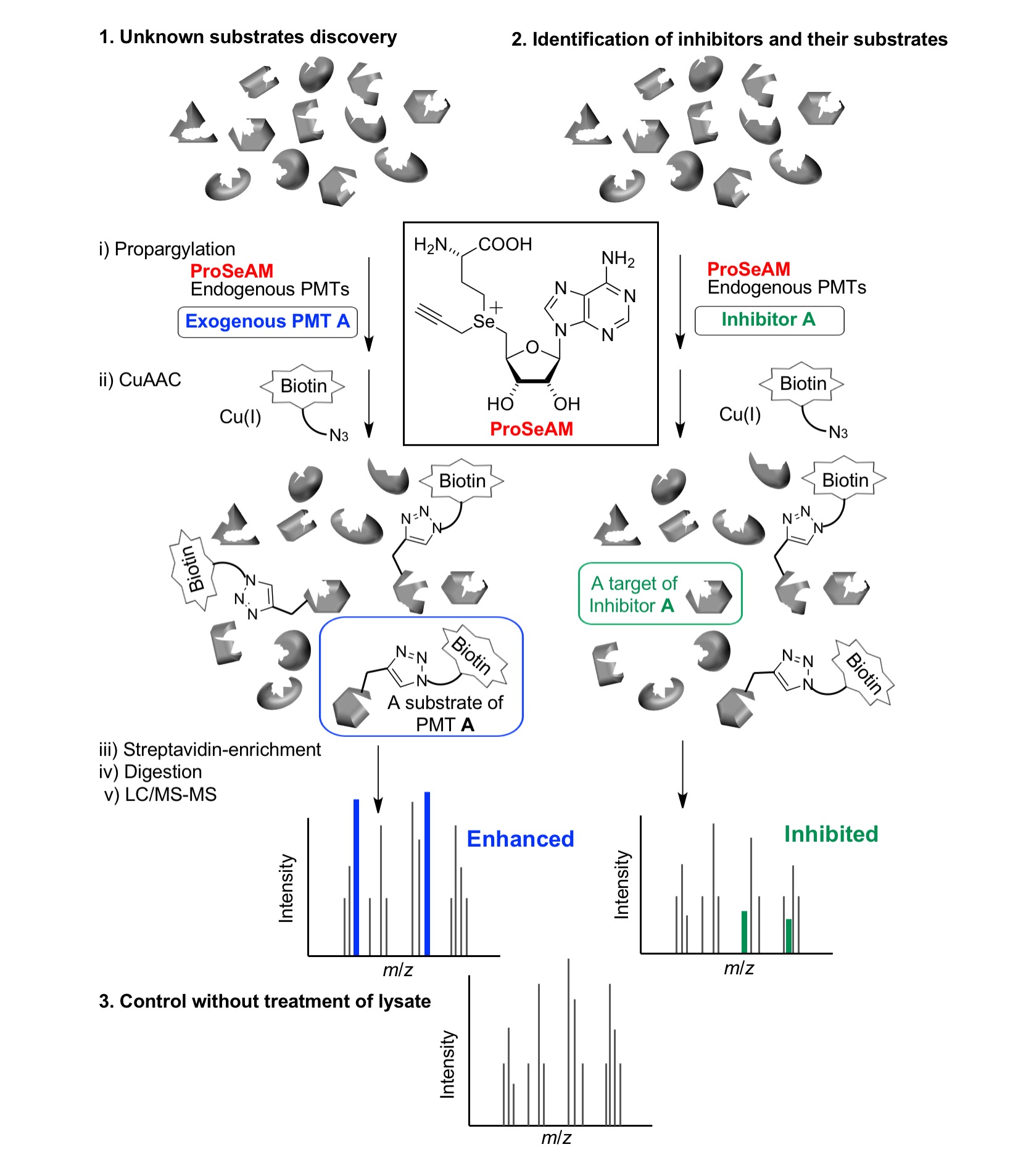
Press release
Development of Indolylmaleimide (IM) derivatives as a novel cell death inhibitor and their therapeutic application for ischemia-reperfusion injury model.
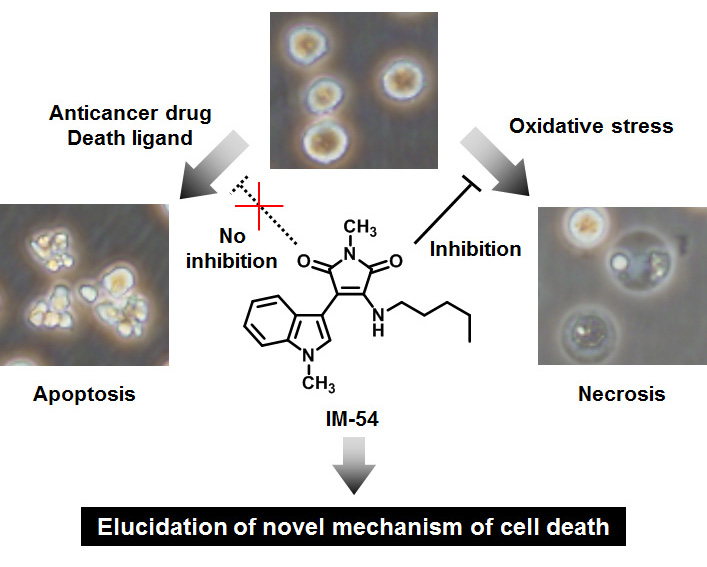
Cell death is one of the most important events for maintaining the health of multicellular organisms. In general, cell death is classified into two major categories, apoptosis and necrosis, according to its morphological features. Apoptosis is associated with characteristic and regulated morphological changes, such as membrane blebbing, cellular shrinkage, nuclear condensation, and formation of apoptotic bodies. In contrast, necrosis results in cellular swelling and rupture of cell membrane, which could be induced by accidental physical injury. Therefore, only apoptosis was believed to be regulated, and necrosis had been considered as uncontrolled and accidental cell death. However, recent studies have revealed novel signaling pathways for some types of necrosis. In addition, necrosis was reported to be involved in some diseases, such as ischemia-reperfusion injury, but its physiological role and detailed mechanism remain to be clarified. Therefore, necrosis inhibitors are expected to contribute the biological studies of necrosis.
We previously developed indolylmaleimide (IM) derivatives as novel inhibitors for necrosis induced by oxidative stress. IM-54, the most potent derivative, did not inhibit apoptosis induced by physiological death ligands or anticancer drugs, indicating the selectivity for necrosis.
To examine the involvement of necrosis inhibited by IM-54 in diseases, we planned to apply IM derivatives for disease models. Due to the low water-solubility of IM-54, we performed structural development of IM derivatives suitable for in vivo studies. As a result, IM-17 was obtained as a water-soluble derivative having comparable activity to IM-54. Furthermore, IM-17 showed higher stability against liver metabolism than IM-54. Taken together, IM-17 was applied for ischemia-reperfusion injury model, in which oxidative-stress-induced necrosis is thought to play an important role. As a result, IM-17 decreased mortality from 60 % to 0% and perfectly prevented sudden death in ischemia-reperfusion-induced arrhythmia model. These results indicate the therapeutic potential of IM derivatives for ischemia-reperfusion injury.
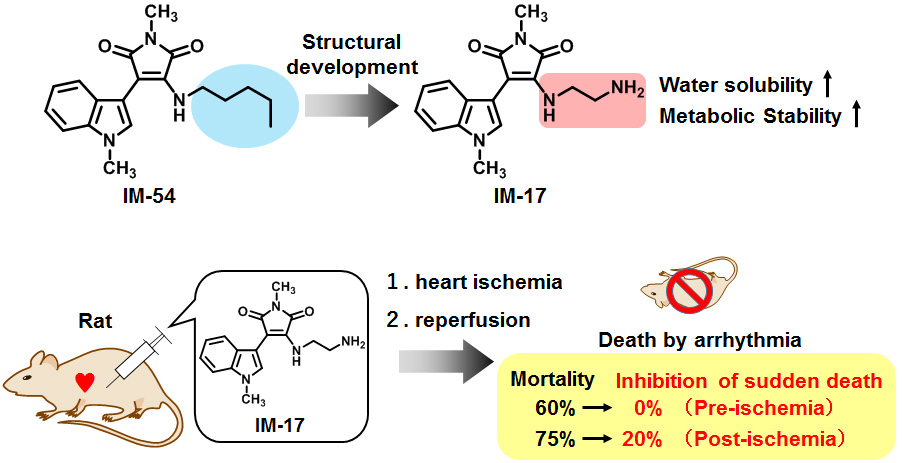
Press release
4. Alkyne-tag Raman Imaging (ATRI) ~Visualization of small molecules in live cells~
To analyze the cellular localization of proteins or compounds in cells, fluorescent tag is usually introduced into target molecules and visualized by fluorescent microscope. For small molecules, however, fluorescent tag tends to decrease their biological activities due to the bulkiness of fluorescent group. To overcome the problem, we developed imaging method for small molecules by using Raman microscopy (collaboration with Prof. Fujita in Osaka university).
Raman microscopy detects specific vibrational signals of molecules based on Raman scattering. Alkyne is a small functional group and shows an intense Raman signal in a cellular Raman-silent region that is free of interference from endogenous molecules. Therefore, alkyne tag was expected to be selectively detected by Raman microscopy without affecting the biological activities of small molecules. To demonstrate alkyne-tag Raman imaging (ATRI), the imaging of EdU (5-ethynyl-2’-deoxyuridine), a DNA synthesis probe, was examined. As a result, EdU was successfully detected in living HeLa cells. This is the first example of Raman imaging of alkyne-tagged molecules in living cells.
We applied ATRI for the visualization of raft structures in artificial lipid membrane (collaboration with Prof. Murata in Osaka University), the mechanistic study of plant pathogen coronatine (collaboration with Prof. Ueda in Tohoku University), and the identification of binding proteins/sites of bioactive compounds. We are now applying ATRI for mechanistic studies of various bioactive compounds.
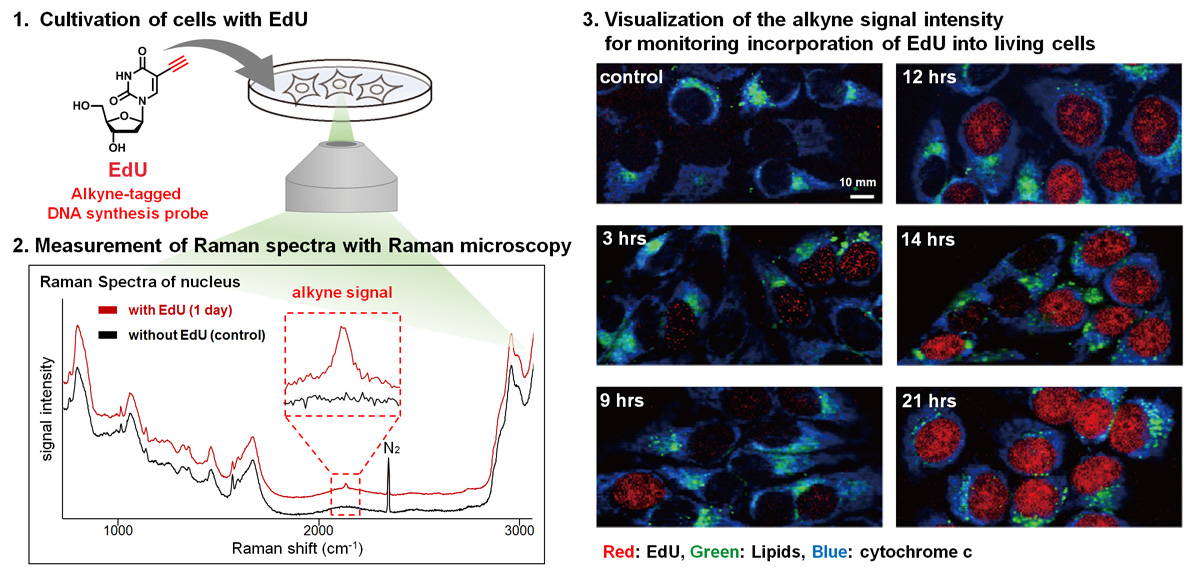
Press release
- Tohoku Univ. press release (May 17, 2017)
- RIKEN RESEARCH Research Highlights (July 17, 2015)
- RIKEN RESEARCH Research Highlights (April 12, 2013)
- RIKEN NEWS (June, 2011)
- RIKEN RESEARCH Research Highlights (May 27, 2011)
- JST press release (March 22, 2011)
5. Methodologies for identification of binding proteins/sites of bioactive compounds.
In chemical biology research, target identification of bioactive compounds is important for mechanistic study, but there are various difficulties. To overcome this problem, we develop methodologies to identify binding proteins/sites of bioactive compounds by using new chemical tags or units.
1)Turn-ON fluorescent Affinity Labeling: O-NBD method
Fluorescent affinity labeling has become a powerful tool to identify and visualize binding proteins of bioactive compounds in live cells. In general, fluorescent group and reactive group are introduced into target compound. However, although various functional groups have been utilized, two functional groups often decrease biological activities of target compound.
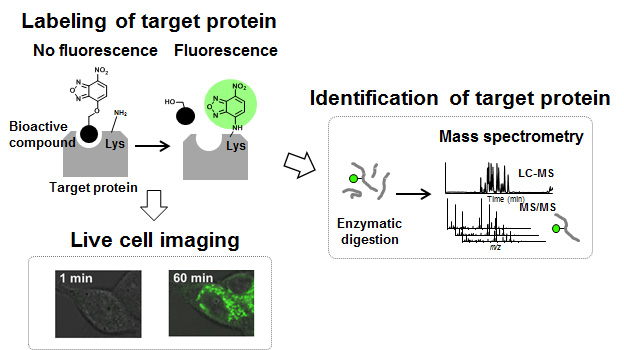
To address this problem, we designed a small bifunctional O-NBD unit (NBD: nitrobenzoxadiazole), which can work as both fluorescent and reactive groups. The O-NBD unit itself is non-fluorescence, but it can react with amine (lysine) and turn into N-NBD unit, which exhibits a strong fluorescence. Therefore, the O-NBD unit is expected to label lysine residues of binding proteins with “turn-ON” fluorescence. Moreover, because of its small size (MW: 180), the O-NBD unit is expected to have little effect on the ligand-protein interaction and biological activities of original compounds. After labeling, fluorescence detection enables us to identify binding proteins/sites of target proteins.
Press release
2) Alkyne-Tag Raman Screening (ATRaS)
We already demonstrated alkyne-tag Raman imaging (ATRI) for analyzing cellular distribution of small molecules in live cells. We also applied Raman detection for identification of binding proteins/sites of bioactive compounds and developed alkyne-tag Raman screening (ATRaS).
In general, to determine the binding sites, target proteins are labeled with bioactive compounds and digested into peptide mixture, and then labeled peptides are identified by LC-MS/MS analysis. However, large amounts of unmodified peptides often disturb the detection of labeled peptides by mass spectroscopic analysis. To overcome this problem, we planned ATRaS, in which alkyne-tagged probes are used to label target proteins and labeled peptides are detected by quantitative Raman analysis. To develop the ATRaS system, we selected cathepsin B, one of lysosomal proteases, as a model protein and its covalent inhibitor Z-FG-AOMK. To realize Raman detection of alkyne-tagged peptides, we found that Surface-enhanced Raman Scattering (SERS) using silver nanoparticles dramatically improved the detection limit and finally succeeded in identifying the binding site of Z-FG-AOMK with ATRaS system.
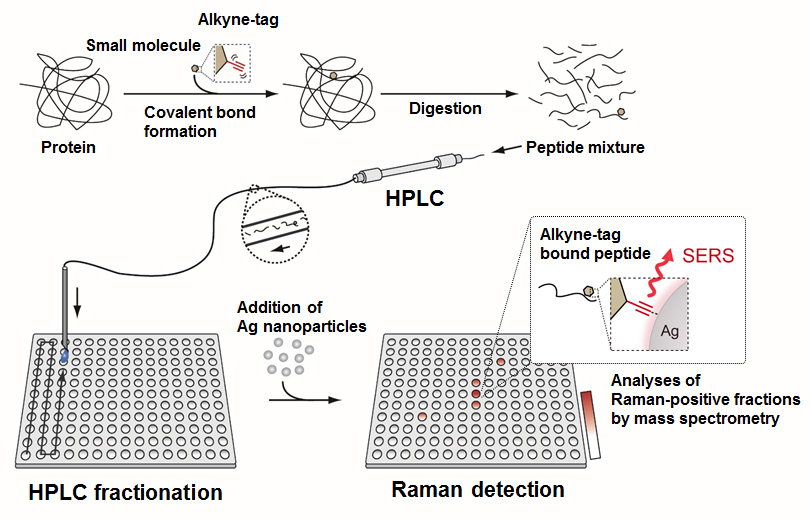
In addition, to improve the efficiency of ATRaS system, we also developed a novel method to purify alkyne-tagged molecules by using cobalt complex. We will apply these methods for target identification of bioactive compounds.